Commits on Source (95)
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
GURMAIL SINGH authored
-
GURMAIL SINGH authored
-
GURMAIL SINGH authored
-
GURMAIL SINGH authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
- /Labs/Lab9/EDGAR.md - /Labs/Lab9/README.md
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
JINLANG WANG authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
gsingh58 authored
-
JINLANG WANG authored
-
gsingh58 authored
Showing
- Labs/Lab1/lab1.md 1 addition, 1 deletionLabs/Lab1/lab1.md
- Labs/Lab1/vm/gcp/gcp.md 1 addition, 1 deletionLabs/Lab1/vm/gcp/gcp.md
- Labs/Lab10/README.md 7 additions, 0 deletionsLabs/Lab10/README.md
- Labs/Lab11/README.md 9 additions, 0 deletionsLabs/Lab11/README.md
- Labs/Lab11/SQL.md 156 additions, 0 deletionsLabs/Lab11/SQL.md
- Labs/Lab11/counting-cells.md 49 additions, 0 deletionsLabs/Lab11/counting-cells.md
- Labs/Lab11/raster.md 93 additions, 0 deletionsLabs/Lab11/raster.md
- Labs/Lab12/8-geo/city.zip 0 additions, 0 deletionsLabs/Lab12/8-geo/city.zip
- Labs/Lab12/8-geo/expected.png 0 additions, 0 deletionsLabs/Lab12/8-geo/expected.png
- Labs/Lab12/8-geo/lakes.zip 0 additions, 0 deletionsLabs/Lab12/8-geo/lakes.zip
- Labs/Lab12/8-geo/main.ipynb 282 additions, 0 deletionsLabs/Lab12/8-geo/main.ipynb
- Labs/Lab12/8-geo/solution.ipynb 225 additions, 0 deletionsLabs/Lab12/8-geo/solution.ipynb
- Labs/Lab12/8-geo/street.zip 0 additions, 0 deletionsLabs/Lab12/8-geo/street.zip
- Labs/Lab12/README.md 17 additions, 0 deletionsLabs/Lab12/README.md
- Labs/Lab12/dot-product-matrix-multiplication/README.md 106 additions, 0 deletionsLabs/Lab12/dot-product-matrix-multiplication/README.md
- Labs/Lab12/model-comparison/README.md 139 additions, 0 deletionsLabs/Lab12/model-comparison/README.md
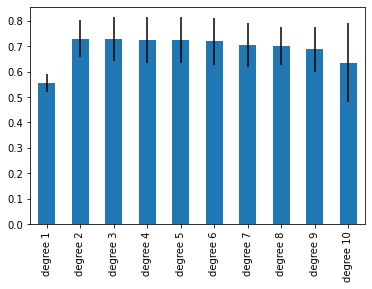
- Labs/Lab12/model-comparison/compare.png 0 additions, 0 deletionsLabs/Lab12/model-comparison/compare.png
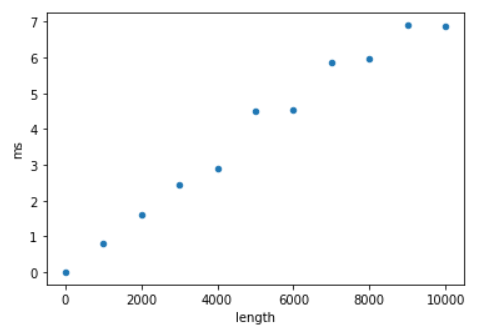
- Labs/Lab12/model-comparison/data.png 0 additions, 0 deletionsLabs/Lab12/model-comparison/data.png
- Labs/Lab12/regression/README.md 69 additions, 0 deletionsLabs/Lab12/regression/README.md
- Labs/Lab12/regression/regression.png 0 additions, 0 deletionsLabs/Lab12/regression/regression.png
Labs/Lab10/README.md
0 → 100644
Labs/Lab11/README.md
0 → 100644
Labs/Lab11/SQL.md
0 → 100644
Labs/Lab11/counting-cells.md
0 → 100644
Labs/Lab11/raster.md
0 → 100644
Labs/Lab12/8-geo/city.zip
0 → 100644
File added
Labs/Lab12/8-geo/expected.png
0 → 100644
232 KiB
Labs/Lab12/8-geo/lakes.zip
0 → 100644
File added
Labs/Lab12/8-geo/main.ipynb
0 → 100644
Source diff could not be displayed: it is too large. Options to address this: view the blob.
Labs/Lab12/8-geo/solution.ipynb
0 → 100644
This diff is collapsed.
Labs/Lab12/8-geo/street.zip
0 → 100644
File added
Labs/Lab12/README.md
0 → 100644
Labs/Lab12/model-comparison/README.md
0 → 100644
Labs/Lab12/model-comparison/compare.png
0 → 100644
6.2 KiB
Labs/Lab12/model-comparison/data.png
0 → 100644
7.66 KiB
Labs/Lab12/regression/README.md
0 → 100644
Labs/Lab12/regression/regression.png
0 → 100644
19.2 KiB